
Option to a large value to try to improve finding matches in repetitive regions and do not When using the command line version of BLAT, you can set the repMatch See Downloading BLAT source and documentation for 2bit file of a genome to your own machine and use BLAT on
Alignment tool for mac download#
If this is not possible, the only alternative is to download You can work around this minimum length limitation but adding more flanking sequence to your query You canĬheck whether any sequence is present at a particular location Often for repeat sequences, you can use theįind the other matches, but only if the other matches are long and specific enough. This is done to improve speed, but can result in missed hits when you Genome and limits results to 16 matches per chromosome strand. The online version of BLAT masks 11mers from the query that occur more than 1024 times in the Leading to missing matches for these repeat sequences. The In-Silico PCR tool is more sensitive and should be preferred for pairs of primers.Īnother problematic case is searching for sequences in repeats or transposons.īLAT skips the most repetitive parts of the query and limits the number of matches it finds,
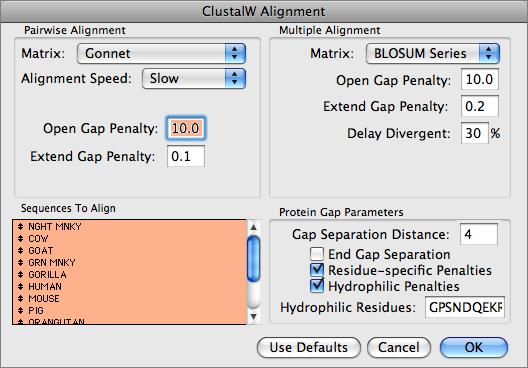
In-Silico PCR and selecting a gene set as the target. Very short sequences that go over a splice site in a cDNA sequence can't be found, as they are not Many published articles do not specify the assembly version so trying both may be necessary. Human genome are currently in wide use (hg19 and hg38) and your sequence may be only in one of Am I doingįirst, check if you are using the correct version of the genome. BLAT can't find a sequence or not all expected matches I can't find a sequence with BLAT although I'm sure it is in the genome. Sensitivity changes by installing command-line Blat on their own Linux server. Structure of an mRNA, but expert users can run large batch jobs and make internal parameter The ability to submit a long list of simultaneous queries in fasta formatĪlignment block details in natural genomic orderĪn option to launch the alignment later as part of a custom trackīLAT is commonly used to look up the location of a sequence in the genome or determine the exon Speed (no queues, response in seconds) at the price of lesser homology depth Psi-BLAST at NCBI can find much more remote matches.įrom a practical standpoint, BLAT has several advantages over BLAST: Within humans, proteinīlat gives a much better picture of gene families (paralogs) than DNA Blat. Terrestrial vertebrates and even earlier organisms for conserved proteins. In practice - due to sequence divergence rates over evolutionary time - DNAīLAT works well within humans and primates, while protein Blat continues to find good matches within The protein index requires slightly more thanĢ gigabytes of RAM. Similarity to the query of length 20+ amino acids. On proteins, BLAT uses 4-mers rather than 11-mers, finding protein sequences of 80% and greater Slightly different from those on the graphical version of BLAT.) (The default settings and expected behavior of standalone Blat are Sequences of 95% and greater similarity of length 40 bases or more. This smaller size means that BLAT is far more easily
The index consists of all non-overlapping 11-mers except for those heavily involved in repeats, and GenBank sequences, but instead an index derived from the assembly of the entire genome. Thus, the target database of BLAT is not a set of Keeping an index of an entire genome in memory. BLAST What are the differences between BLAT and BLAST?īLAT is an alignment tool like BLAST, but it is structured differently. Protein-translated BLAT having different results.Standalone or gfServer/gfClient result start positions off by one.Approximating web-based BLAT results using gfServer/gfClient.Using BLAT for short sequences with maximum sensitivity.Replicating web-based BLAT "I'm feeling lucky" search.Replicating web-based BLAT percent identity and score calculations.Replicating web-based BLAT parameters in command-line version.Downloading BLAT source and documentation.BLAT or In-Silico PCR finds multiple matches such as chr_alt or chr_fix even though only one is.BLAT cannot find a sequence at all or not all expected matches.


 0 kommentar(er)
0 kommentar(er)
